Help/Tutorial
CancerVar's help page can be found here.
Step by Step
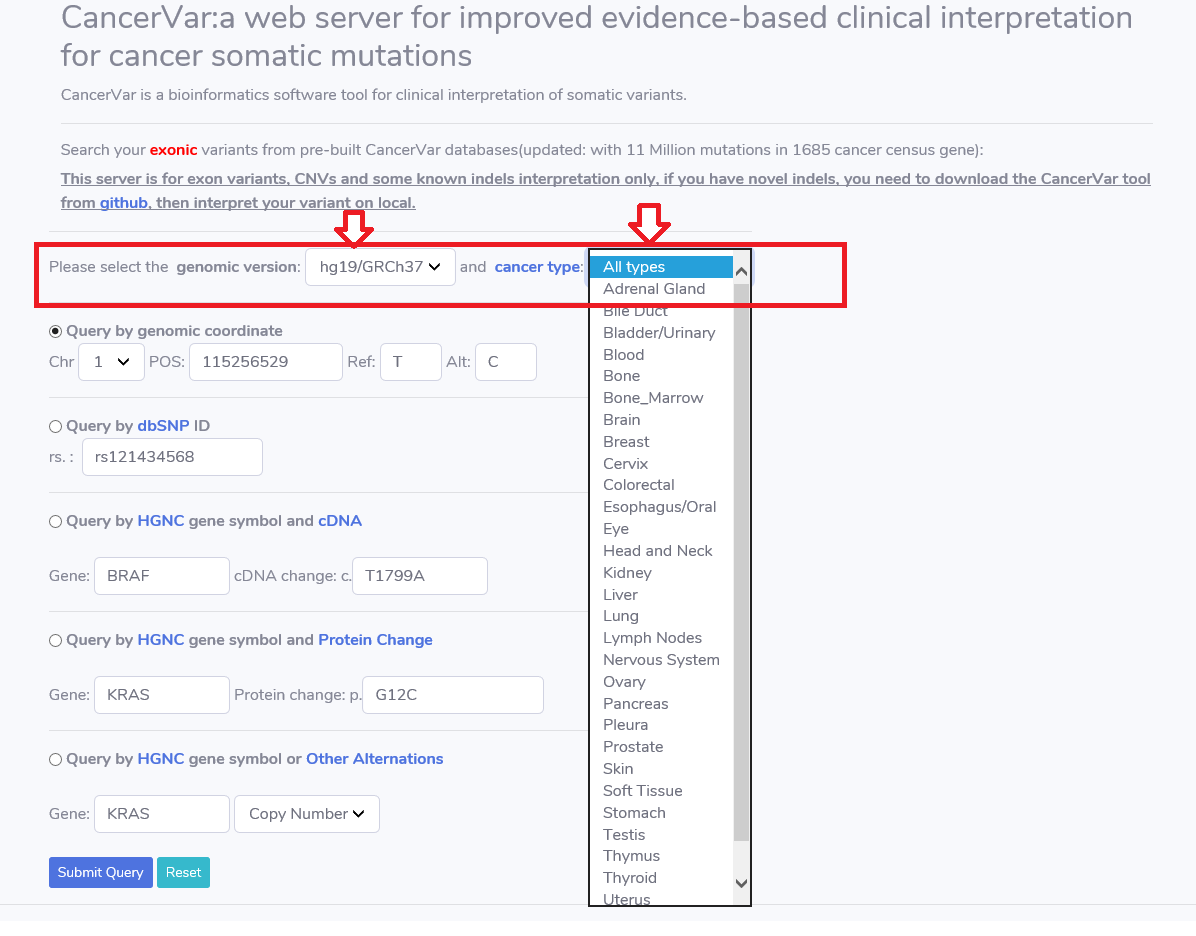
1.Choose the genomic version and proper cancer type.
Before the variant searching, the user need to select the genomic version(currently, support hg19 and hg38) and cancer type.
CancerVar group the cancers into 30 types based on the primary site.If user dose not sure the cancer type, then choose the types as "All types".
2.Choose the searching method
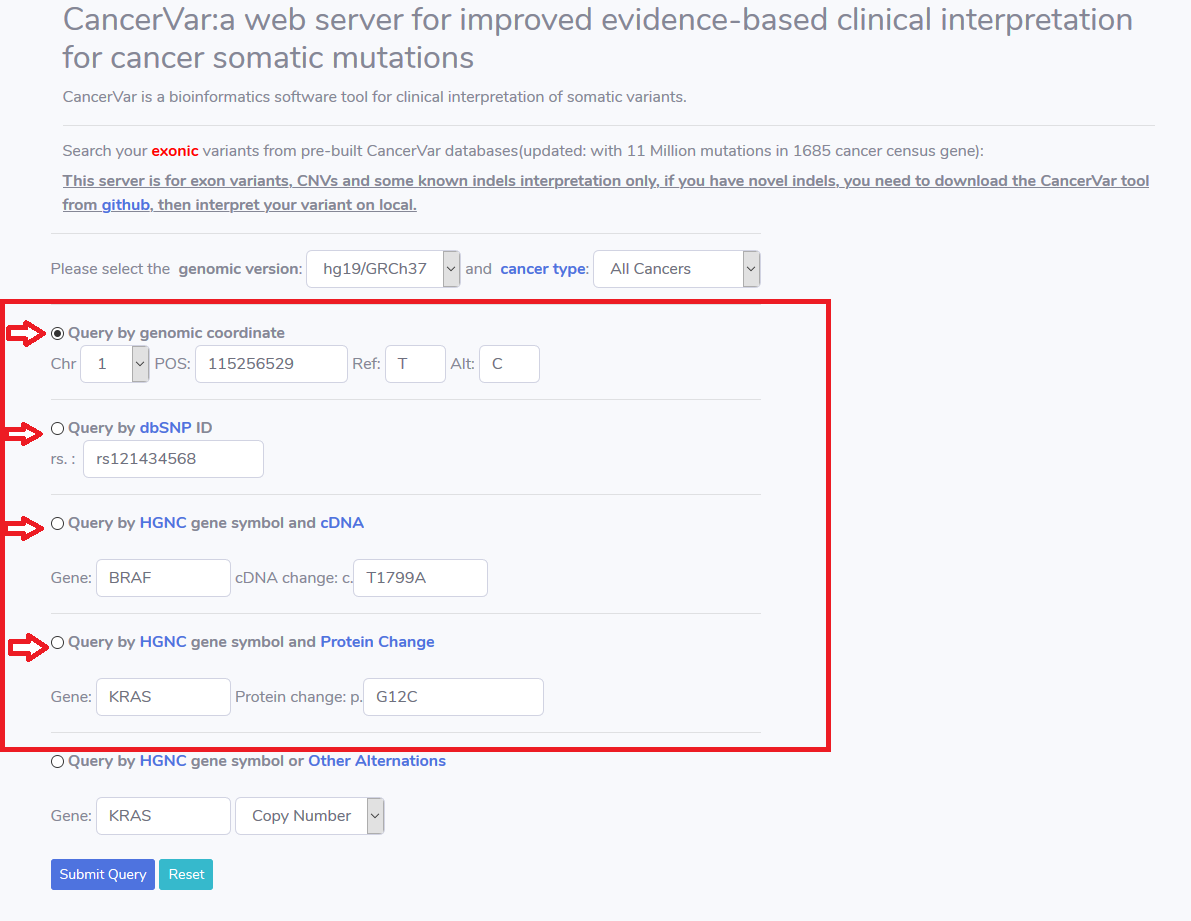
When searching Non Structural variation(SNV,indel), user can choose using :
2.a) genomic coordinate with reference and alternative allele.
2.b) dbSNP with rs#.
2.c) HGNC gene symbol and cDNA change.
2.d) HGNC gene symbol and protein change.
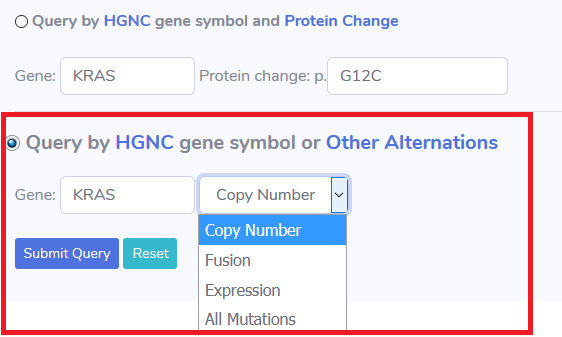
When searching other alternations(such as CNV, fusion, expression), user can choose :
2.e) HGNC gene symbol and Copy number variation.
2.f) HGNC gene symbol and fusion
2.g) HGNC gene symbol and expression.
2.h) all the alternation in this HGNC gene symbol .
3.Click submit to get the result.
Submit the query.
4.Review the interpretation result.
4.1 Interpretation summary.
In this card, CancerVar will give the interpretation result as Pathogenic,Likely Pathogenic, Unknown significance or Benign/Likely Benign.
CancerVar summarized the evidence used for the interpretation of clinical significance.
Followed by three cards: Gene Overview , Mutation Information and Evidence Overview.
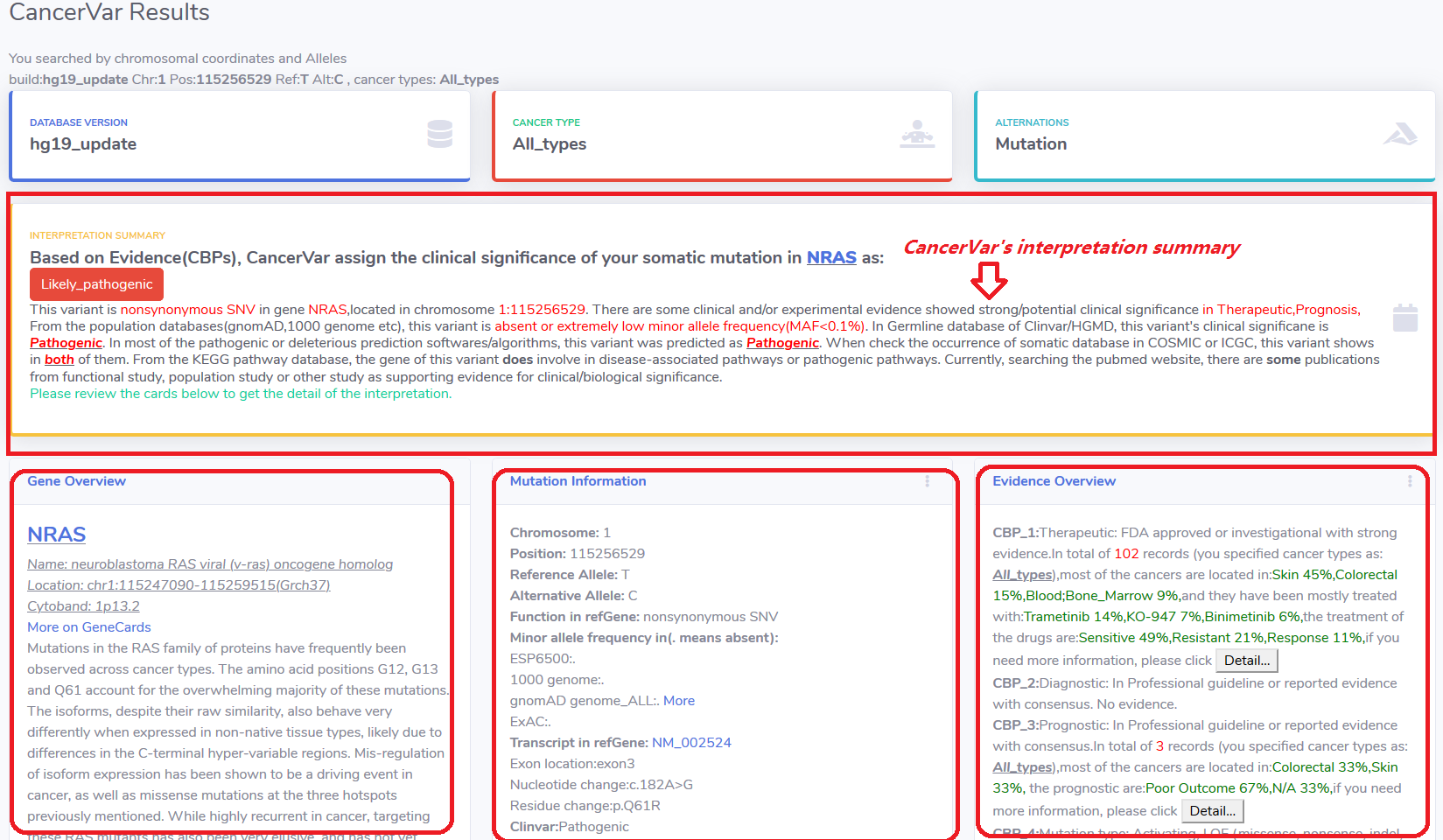
4.2 Gene Overview.
This card provide gene's description related with cancer.
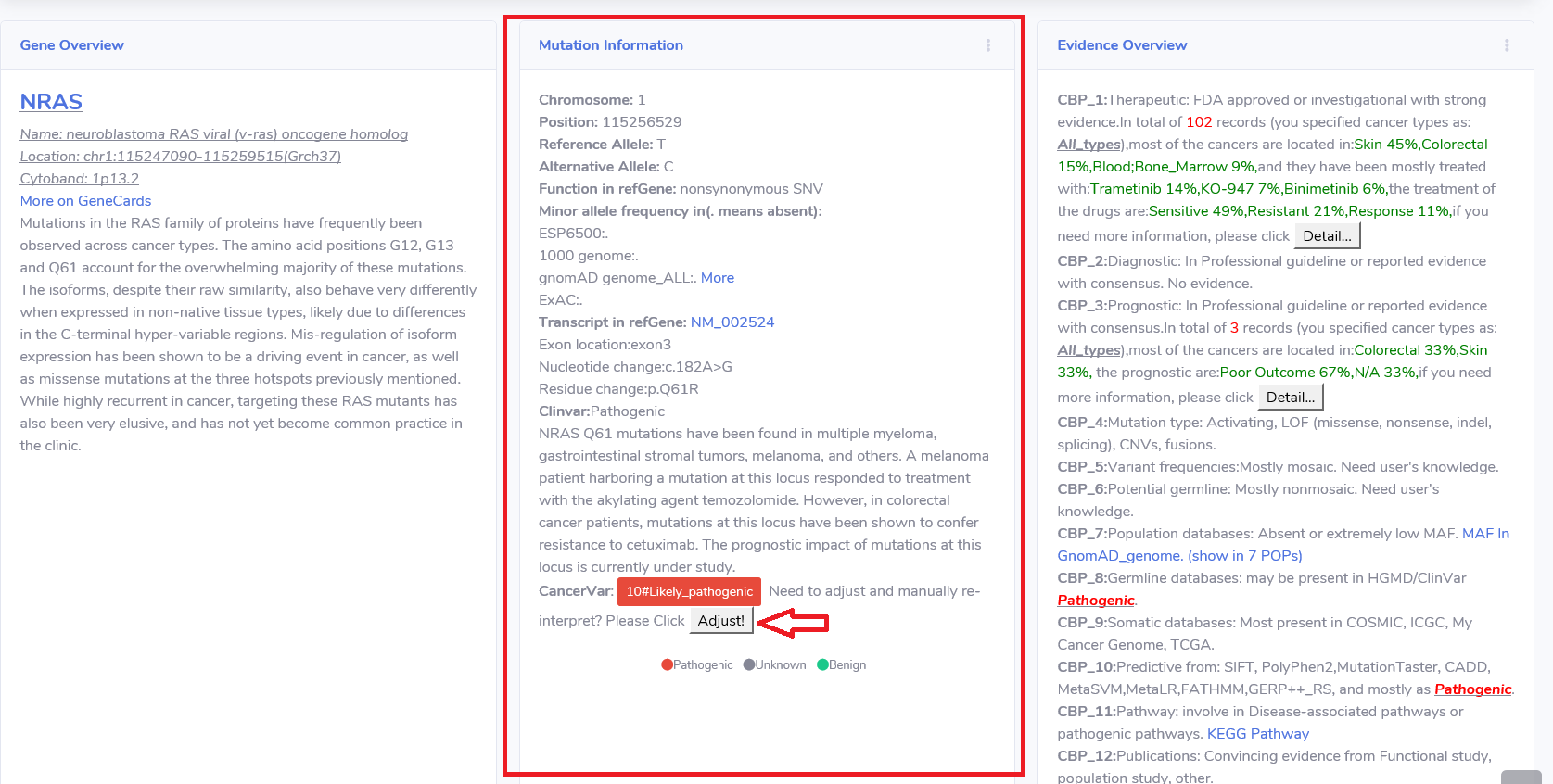
4.3 Mutation Information.
This card provide some genomic information of this variant, also some description related with cancer.
The CancerVar's interpretation also showed here as "score#clinical significance".
If you have more information or knowledge, you can click the "Adjust!" button to re-interpret the variant.
4.4 Evidence Overview.
This card list all the evidence for the interpretation.
If there are no evidence to support the interpretation, we will note as "No evidence" after the evidence description.
For CBP1 as therapeutic evidence, CancerVar provide the drug response and related cancer information.
For CBP2 as diagnostic evidence, CancerVar provide diagnostic significance(Positive/Sensitive,Negative).
For CBP3 as prognostic evidence, provide the outcome(poor or better) information.
If user want to explorer more such as publications,then click the "detail" button.
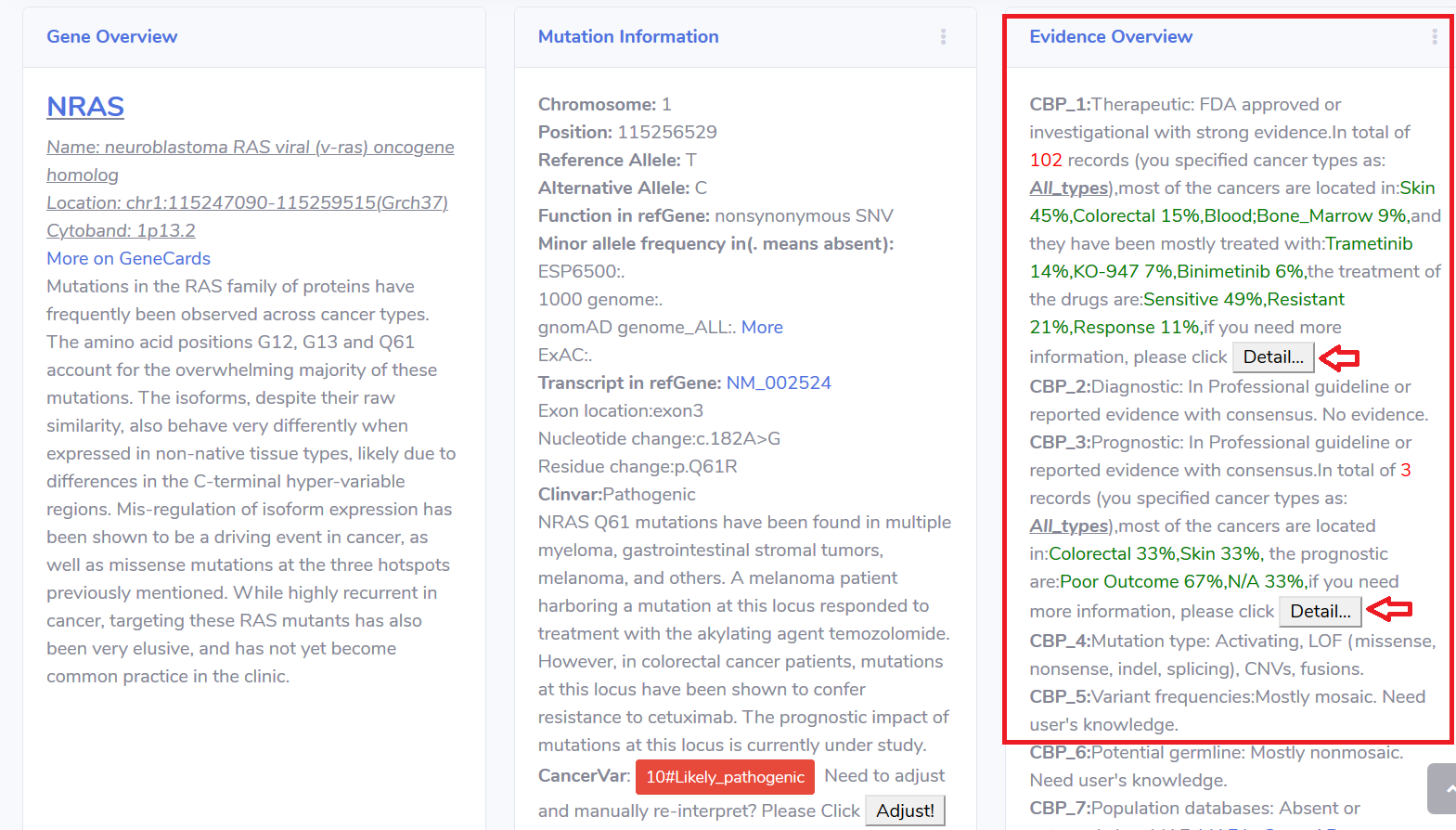
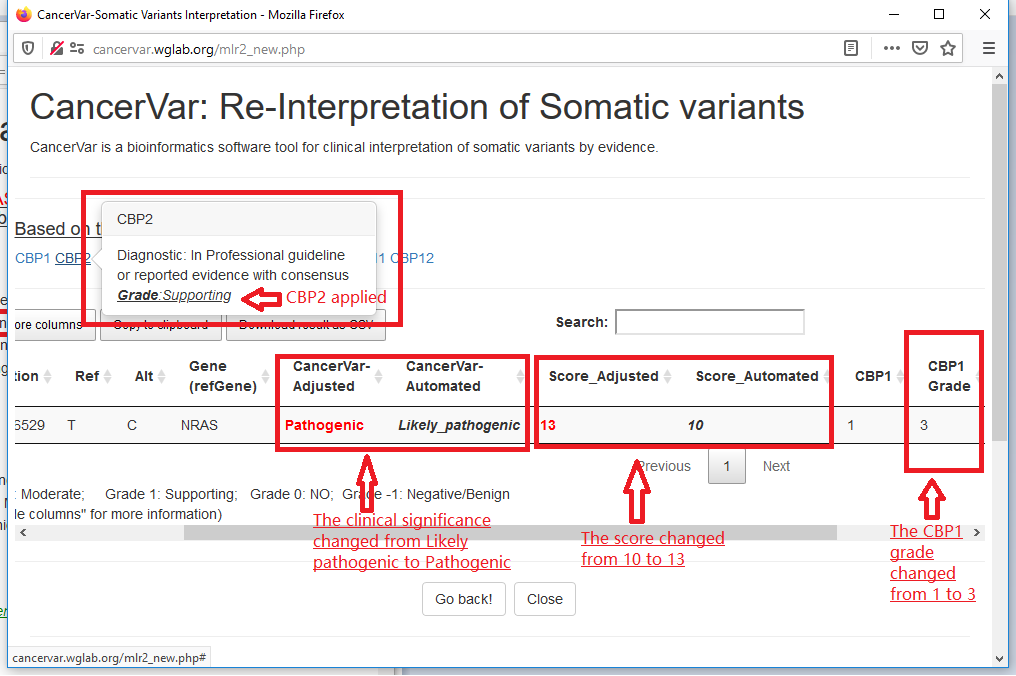
5.Re-Interpret or Manually adjust the interpretation result based on user's experience.
5.1 In the "Mutation Information" card, click the button "Adjust!" and wait to popout the page.
5.2 check or uncheck evidence and re-select grade in the re-interpretation page.
In this sample, the variant has no evidence for CBP2(Diagnostic),if we want to apply this evidence, we check "CBP2" and select the supporting grade.
We also upgrade the evidence of "CBP1" from "supporting" to "strong".
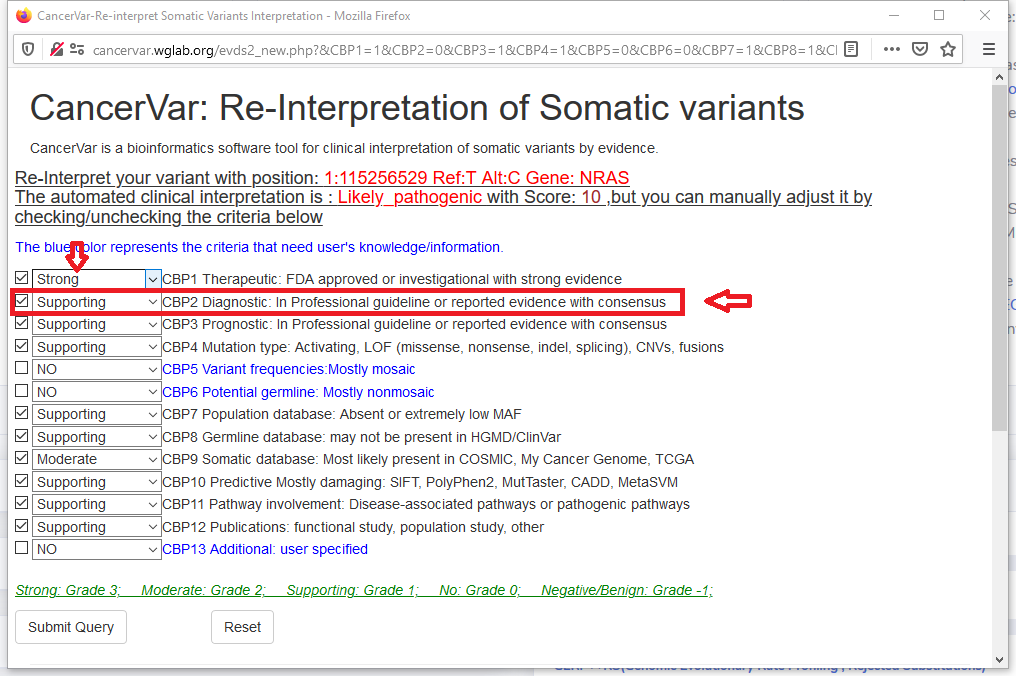
5.3 Submit, we will get the new interpretation: